VMD
VMD Modular Sorter Cost: 500L$ This is the heart of my system. You can sort all of your DFS products here and store them for later use. This comes with everything you need to get started with your cooking. You will be able to create any crate you want for your sorters. VMD Ethics Hotline Tel: (844) 415-9857. Corporate Headquarters. 4114 Legato Road #700, Fairfax, VA 22033 (571) 612-2424. Airport Services (800) 908-8110. Atlantic City, NJ. Kansas City, MO. Type and Press “enter” to Search.
- VMD is designed for modeling, visualization, and analysis of biological systems such as proteins, nucleic acids, lipid bilayer assemblies, etc. It may be used to view more general molecules, as VMD can read standard Protein Data Bank (PDB) files and display the contained structure.
- The Veterinary Medicines Directorate (VMD) protects animal health, public health and the environment. VMD is an executive agency, sponsored by the Department for Environment, Food & Rural Affairs.
Return to Main Documents Page
This is a very basic introduction to VMD.
So, what is VMD? In the words of its developers:
VMD (Visual Molecular Dynamics) is a molecular visualization and analysis program designed for biological systems such as proteins, nucleic acids, lipid bilayer assemblies, etc.It is developed by the Theoretical and Computational Biophysics Group at the University of Illinois at Urbana-Champaign.Among molecular graphics programs, VMD is unique in its ability to efficiently operate on multi-gigabyte molecular dynamics trajectories, its interoperability with a large number of molecular dynamics simulation packages, and its integration of structure and sequence information.
The aim of this tutorial is to very quickly get you familiar enough with VMD to be able to view individual protein structures and the sorts of trajectories containing many structures that are produced by molecular dynamics and other simulation techniques.This document is deliberately designed to cover only the most basic features of VMD.Excellent tutorials teaching the full range of the functionality provided by the program can be found at the VMD website, here.
Prerequisites
This tutorial assumes three things:
- Access to the following two files (download and save them in a location of your choice):
- a PDB single structure file, dbd.pdb
- a DCD multiple structure trajectory file, dbd-short-traj.dcd
- VMD is installed on your computer.
- You have started VMD.
If you don't currently meet the last two criteria then follow the instructions in the next two sections.
Installing VMD
If VMD is not already available on your computer then you will, obviously, need to install it.Fortunately, VMD is free and pre-compiled ready to go versions are available for Windows, OS X and Linux.You just need to register with your email address and then download the appropriate package from here.
Full installation instructions are available here, but the Windows and OS X installations are pretty self explanatory.The small extra effort involved in Linux installation is also documented in the README file distributed with the program.
Running VMD
To start the program:
- OS X: Double click on the VMD icon in the Applications directory.
- Linux: Type vmd in a terminal window.
- Windows: Select Start ->Programs ->VMD.
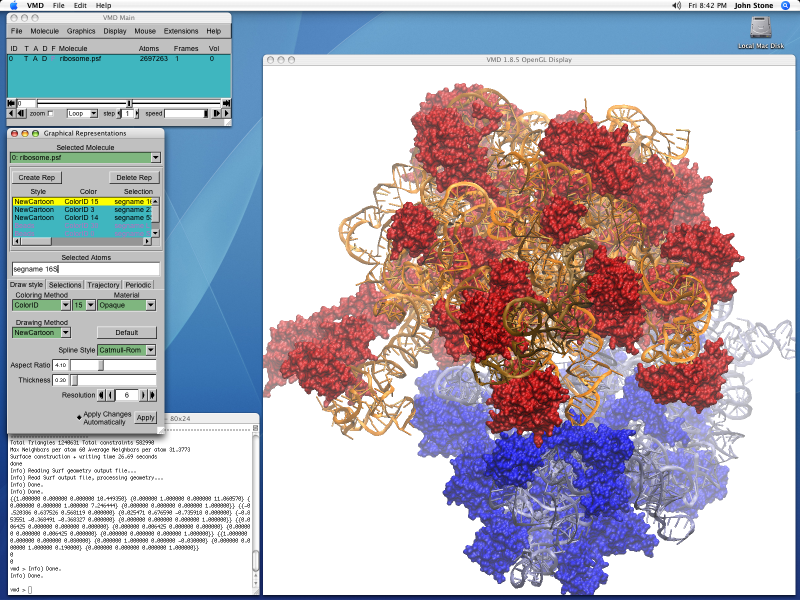
Upon opening VMD opens three windows (Figure 1): the Main, OpenGL Display and Console (or Terminal on OS X) windows.To end a VMD session, go to the Main window, and choose File ->Quit.You can also quit VMD by closing the Console or Main window.
Viewing Single Molecules
In this section we will load a single structure from a PDB and learn how to view it from different angles and to alter the way it is rendered on screen.
Loading a Molecule
- From the menubar in the Main window select File ->New Molecule... (Figure 2).
- A Molecule File Browser window similar to that shown in Figure 3 should now open.Select the Browse.. button (circled in red in Figure 3).This will open a file selection dialog.Navigate to the PDB file, dbd.pdb, which you downloaded earlier, select it and click Open.
- You should now have been returned to the Molecule File Browser window (the structure will not yet have been loaded).To load the file you need to click the Load button (circled in blue in Figure 3).The structure should now be loaded and the OpenGL Display window look something like Figure 4.
Notice that once the molecule is loaded basic information including the name of the file and number of atoms appear in the Main window.
Once the structure is loaded you can close the Molecule File Browser at any time.
Altering Your Viewpoint
From the menubar in the Main window select Mouse (Figure 5).
A larger array of options for how the mouse interacts with the molecule shown in the OpenGL Display.We will concentrate on the first four options: rotate mode, translate mode, scale mode and center.The keyboard shortcut for each option is shown next to the choice in the menu (for example pressing 't' when the OpenGL windows is selected will change into translate mode).By default VMD uses the rotate mode.
Vmd Drogerie
Generally, to change the viewpoint you need select the OpenGL Display windows, hold the left mouse button and move the mouse (the right mouse button can be used in rotate mode, see below).Have a play with all of the modes below until you feel comfortable changing the view and switching between the various modes.You can easily tell which mode you are in because each one has a distinctive cursor (Figure 6).
Rotate mode: With the left mouse button held moving the mouse horizontally rotates the molecule around the vertical axis, up and down around a horizontal one.When the right mouse button is held then the rotation is performed around an axis running into and out of (i.e. perpendicular to) the screen.
Translate mode: When you hold down the left mouse button you can now move the molecule up, down, left or right in the viewing plane.
Scale mode: Moving the mouse left or right whilst holding the left mouse button zooms in and out.
Center: The centre option is not a direct method for altering the view point.It is used to pick a point about which to perform a rotation.Once you have selected a center point change to Rotate mode to perform the rotation.
At any point you can reset the view by pressing '=' whilst the OpenGL Display is selected (you can also use the menu option Display ->Reset View.
Changing the Appearance of the Molecule
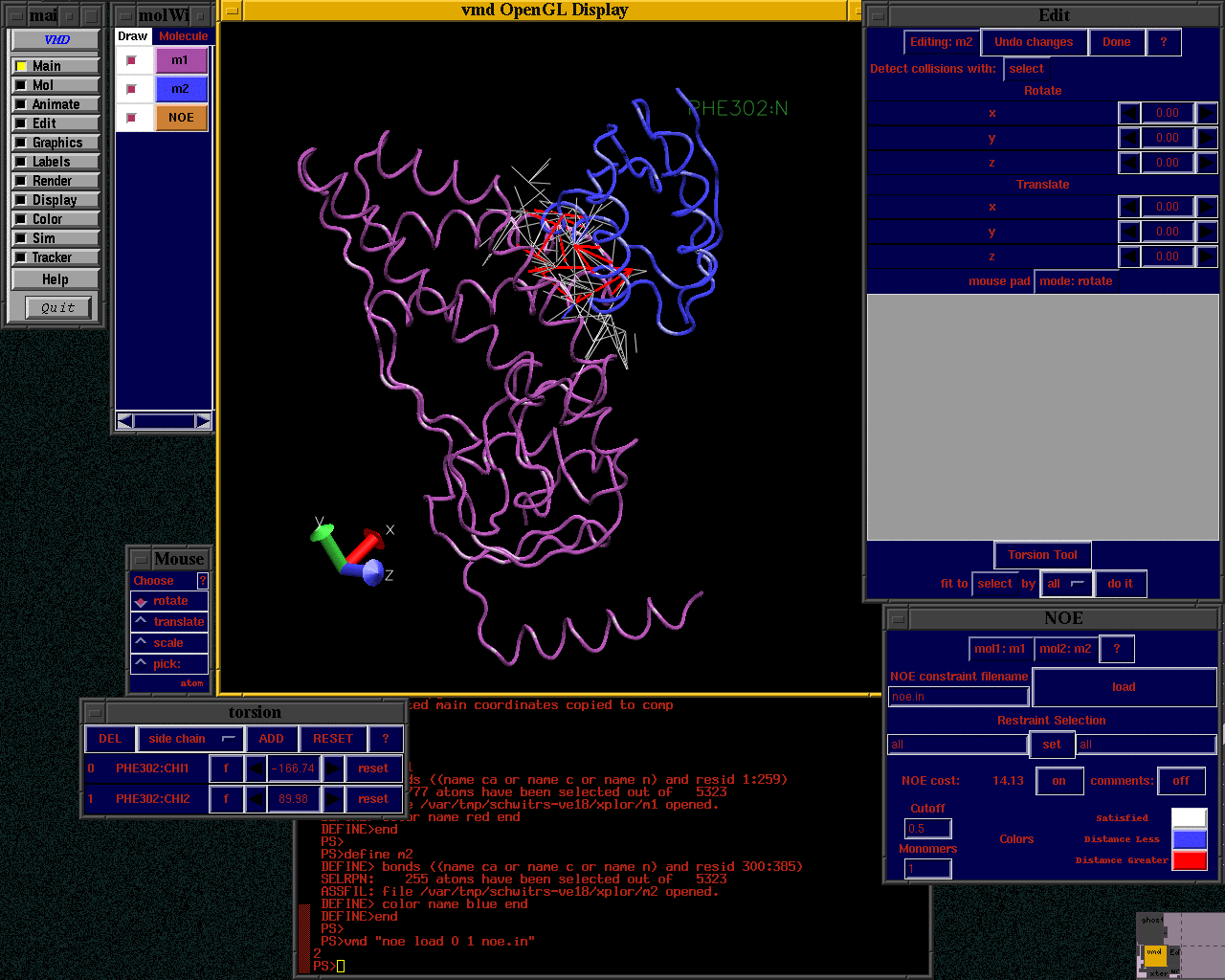
- Select Graphics ->Representations... from the menubar of the Main window.A new Graphical Representations window will open looking like Figure 7.Section Figure 7 (a) shows the current representation being highlighted.It shows the style of the representation (in this case Lines), the colouring method (Name) and the selection of atoms to which this representation is applied.The selection is determined by the contents of the input box shown in Figure 7 (b).The current selection is for all atoms.The language used to select atoms in VMD is documented in the section of the user guide here.
- Try changing the Coloring Method by choosing different options from the drop down menu shown as Figure 7 (c).
- Alter the Drawing Method of the graphics used to display the molecule with the drop down menu shown as Figure 7 (d).A good choice is 'New Cartoon' which highlights different elements of protein secondary structure (shown in Figure 8).Notice how the region indicated by Figure 7 (e) changes to give you different customization options for the different Drawing Methods.
Www.vmd.gov.lv
Working With Trajectories
When dealing with simulations multiple structures are commonly saved together in files called 'trajectories'.An example of a trajectory format is DCD (used by NAMD and CHARMM).Such files contain only coordinates and no information about which atoms are represented.In order to vizualize the atomic structure represented by a trajectory it has to be combined with a file containing structural information.PDB files contain both structural and coordinate information and can perform this role, other filetypes such as PSF files contain only structural information and no coordinates.
The trajectory from this point assumes that you have already loaded the dbd.pdb PDB files into VMD.If that is not the case go through the instructions for [Loading a Molecule].
Loading a Trajectory
In the Main window either select File ->Load Data into Molecule... or right click on the line for dbd.pdb and select Load Data into Molecule... from the menu (see Figure 9).
Like when we loaded a PDB previously a Molecule File Browser window should appear.Browse to the dbd-short-traj.dcd and Load it.
As the trajectory loads you will see the number of frames increase in the Main windows and the structure in the OpenGL Display update.
Vmd Technologies
Trajectory Animation
The Main window should now look something like it does in Figure 10.
However, the current frame (circled in red in Figure 10) should show a number one less than the number of frames shown above it in the information panel.This is because frames are counted from zero in VMD.Frame zero in this case is the structure loaded from the PDB.If you load a structure information only file, like a PSF, instead of a PDB then frame zero will be the first frame of the DCD.
You can use the slider highlighted in blue in Figure 10 to move to any frame you desire.It is also possible to type in a specific frame number in the current frame region.
Beneath these controls are the animation controls (highlighted in green in Figure 10).These allow you to play through the frames of the trajectory.Forwards and backwards play buttons can be found at either end of the controls.Between these are controls which alter the way the animation plays.The 'step' controls the gap between the frames which are displayed and 'speed' is self explanitory.The animation type drop down menu controls what happens when the animation has played the last frame in the current direction:
Vmd Sic
Once: The animation terminates.
Loop: The animation plays again from the start (opposite end).
Rock: The animation reverses direction.
Play with these controls until you are happy you understand how they work.Once you are at that stage our work here is done.
Want to Know More About VMD?
VMD is not only a molecular viewer but also allows you to analyse structures and trajectories, convert file format, render graphics for publication and many more things well beyond the scope of this tutorial.When you want to try out these features best place to start is the User Guide.
When you, inevtiably, run into difficulties a good first port of call is the mailing list.
Vmd Veterinary
Return to Main Documents Page
